Zelluläre Modelle – AG Steenpass

Michaela Hiber, Julia Menges, Theresa Kühnel und Laura Steenpass (vorne)
Projekte
My group works on the development of cellular models to understand the regulation of imprinted gene expression at the Prader-Willi / Angelman syndrome locus and the RB1 gene. We are using human cell lines and human pluripotent stem cells (induced and embryonic). Differentiation of these cells and molecular biology techniques, like genome editing and next generation sequencing, are used for establishment of these models.
Regulation of imprinted gene expression
Genomic imprinting is an epigenetic process regulated by DNA methylation and resulting in parent-of-origin-specific gene expression. DNA methylation is established in only one of the parental germ lines, either the maternal or the paternal one. This results in a methylation mark on only one of the parental chromosomes. Once established, this mark is stable throughout lifetime and in all body cells. But this methylation mark is not read in all cells in the same way.
At the Prader-Willi / Angelman syndrome locus, the maternal chromosome is marked by DNA methylation (Fig. 1). This results in expression of some genes from the paternal allele only in all body cells. The UBE3A gene however, is expressed from both chromosomes in most tissues, except brain. Specifically, in neurons a long non-coding RNA is expressed from the paternal allele, which overlaps the downstream UBE3A gene in antisense direction. This overlap results in competition between the transcription processes of UBE3A and the non-coding RNA leading to silencing of UBE3A gene expression from the paternal allele. If the active chromosome carries a genetic defect, expression of the respective genes will be lost completely and the symptoms of Prader-Willi or Angelman syndrome develop. Prader-Willi syndrome is caused by genetic defects on the chromosome inherited from the father, Angelman syndrome by defects on the chromosome inherited from the mother.
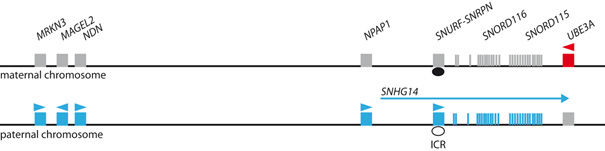
- Fig. 1: Scheme of the Prader-Willi / Angelman syndrome locus on human chromosome 15. Gene expression is regulated by the methylation mark present on the maternal ICR (imprint control region, black ellipse). Genes are expressed only from the paternal allele (blue), because methylation at the ICR is absent (white ellipse). On the maternal chromosome only UBE3A is expressed. UBE3A is silenced on the paternal allele in neurons because of transcriptional interference with SNHG14.
Our aim is to understand the switch from biallelic UBE3A expression to monoallelic expression in neurons of the human brain. For this purpose, we reprogrammed fibroblasts from a patient with Angelman syndrome into induced pluripotent stem cells (iPSCs) and differentiated them into neurons (Fig. 2). Silencing of the paternal allele could indeed be observed in this model. This provides us with an excellent model to study silencing of the paternal copy in an otherwise unchanged genomic environment.
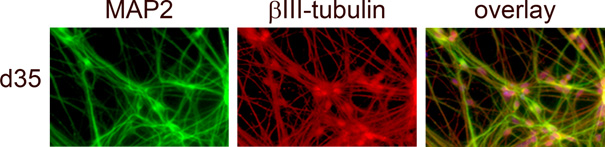
- Fig. 2: Angelman syndrome patient-derived iPSCs differentiated into neurons. MAP2 and ßIII-tubulin protein expression is specific for mature neurons. (Stanurova et al, Scientific Reports 2016).
A second route we follow is the modelling of imprint establishment and transcriptional interference occurring at imprinted gene loci. For this we cloned an upstream inducible promoter in line with a downstream promoter in sense or antisense direction. Using this system in somatic cells, we would like to decipher the factors and circumstances needed for the mentioned regulatory processes.
The imprinted RB1 locus
The second imprinted locus we are working on is the RB1 locus. In humans biallelic inactivation of RB1 results in the development of retinoblastoma. This is a tumor of the developing neural retina occurring in very young children up to the age of five. RB1 imprinted expression is caused by maternal-specific DNA methylation of a retrotransposon that was inserted into intron 2 of the RB1 gene in evolution. The maternal-specific DNA methylation silences an alternative RB1 promoter, named CpG85, on the maternal but not the paternal allele. However, this imprinted DNA methylation mark appears to be unstable and CpG85 is fully methylated in several cell lines and human tissues. It is assumed that paternal-specific expression of the alternative RB1 transcript RB1-E2B reduces transcription from the regular RB1 promoter that is located upstream of CpG85. For an imprinted gene we would expect parent-specific effects on tumor development. Indeed, some rare mutations in RB1 are associated with a higher risk for tumor development when inherited from the father.
The projects dealing with the RB1 locus investigate the prerequisites for stability of the CpG85 imprint and the influence of mutations on the paternal allele on tumor development. To enable these studies, we develop an in vitro system for retinoblastoma, which is based on the differentiation of human embryonic stem cells into neural retina in 3D organoids. Cone photoreceptor precursor cells are the presumed cell-of-origin of retinoblastoma and we could indeed show the presence of these cells in the organoids (Fig. 3). To mimic the RB1 genomic status in heterozygous carriers and the tumor, we introduced RB1 mutations in human embryonic stem cells by CRISPR/Cas9.
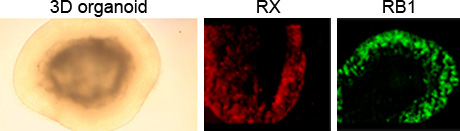
- Fig. 3: Differentiation of human embryonic stem cells into neural retina. Bright outer layer in the organoid represents retinal tissue. In a day 61 organoid, RX stains retinal progenitor cells (red), located in the outer layer. Staining for RB1 protein is shown in green.
Projects and funding
A mouse model for Imprinting of the Human Retinoblastoma Gene
Vasiliki Tasiou, Michaela Hiber, Jana Stanurova
Funding: DFG
Differentiation of human embryonic stem cells into neural retina as in vitro model for retinoblastoma
Laura Steenpass, Deniz Kanber, Gesa Wiel, Julia Menges, Leonie Schipper
Funding: Kinderaugenkrebsstiftung, Deutsches Förderprogramm für Augenheilkunde, Ernst und Bertra Grimmke Stiftung, DFG
Epigenetic mechanisms in Angelman Syndrome: generation and use of patient-derived iPS cells
Jana Stanurova, Anika Neureiter (AG Klump, Institute for Transfusion Medicine, UK Essen)
Funding: IFORES Sonderforschungsprogramm Medical Faculty, University Duisburg-Essen
Modelling transcriptional interference and imprint establishment at the PWS/AS locus in somatic cells
Helena Heinz, Nadja Utz, Theresa Kühnel, Michaela Hiber
Funding: DFG
Members
Laura Steenpass, PI
Deniz Kanber, PostDoc (Prof. Lohmann‘s group)
Anika Neureiter, PhD student (Hannes Klump group, Institute for Transfusion Medicine)
Theresa Kühnel, PhD student
Julia Menges, PhD student
Michaela Hiber, technician
Former members
Nadja Utz, MSc
Vasiliki Tasiou, MSc
Helena Heinz, PhD
Gesa Wiel, MSc
Jana Stanurova, PhD
Leonie Schipper, BSc


